The ABCD_AF291 antibody gives strong and specific signal in ultra-expansion microscopy analysis of HA tagged proteins in African trypanosomes
DOI:
https://doi.org/10.24450/journals/abrep.2024.e1625Keywords:
AF291, epitope, HA, trypanosomeAbstract
African trypanosomes expressing a flagellar central pair protein tagged with a triple hemagglutinin (HA) epitope in tandem repeat were analyzed by ultra-expansion microscopy (U-ExM) using the recombinant anti-HA antibody ABCD_AF291. ABCD_AF291 gave a strong and specific signal, demonstrating that it can be used to localize tagged proteins in U-ExM experiments.
Introduction
The human influenza hemagglutinin (HA) epitope is a short peptide sequence derived from the influenza virus hemagglutinin protein. Because its small size offers minimal functional interference, it is commonly fused to proteins of interest for localisation studies. One such approach is ultrastructure-expansion microscopy (U-ExM), an emerging super-resolution technique that facilitates high resolution protein localisation studies by physically expanding the sample. As with all immunofluorescence-based analyses, however, the quality of the final data is strongly dependent upon the affinity and specificity of the antibody’s interaction with its cognate antigen, and this can vary widely depending on the specific experimental conditions. For this reason, it is important to experimentally validate antibodies for specific protocols prior to their use. Here, we tested the recombinant anti-HA antibody ABCD_AF291 in U-ExM localisation analysis in Trypanosoma brucei, the causative agent of human and animal African trypanosomiasis. We show that ABCD_AF291 is able to detect a triple HA-tagged flagellar central pair protein, PF16 (PF16::3HA) (Smith and Lefebvre, 1996).
Materials & Methods
Antibodies: ABCD_AF291 and ABCD_AA345 (ABCD nomenclature, http://web.expasy.org/abcd/) were produced by the Geneva Antibody Facility (http://unige.ch/medecine/antibodies/) as mini-antibodies with the antigen-binding scFv portion fused to human IgG1 Fc and guinea pig IgG Fc, respectively. The scFv sequence of ABCD_AF291 (AF291) corresponds to the variable regions of the anti-HA monoclonal antibody 12CA5 (Arimo et al., 2017). The scFv sequences of ABCD_AA345 (AA345, anti-tubulin) correspond to the sequences of the variable regions of the clone F2C (Nizak et al., 2003). ScFv variables sequences were joined by the peptide linker (GGGGS)4. Antibodies were produced in HEK293T suspension cells growing in HEK TF medium (Xell #861-0001, Sartorius) supplemented with 0.1% Pluronic F68 (Sigma #P1300). Cells were transiently transfected using FectoPro® transfection reagent (Polyplus #101000014) with the vector coding for the mini-antibodies and supernatants were collected after 3 days.
Antigen:Trypanosoma brucei procyclic SmOx P9 cells (TREU927; Poon et al., 2012) were genetically modified using PCR-tagging (Dean et al., 2015; Paterou et al., 2023) to express the flagellar protein PF16 (UniProt #Q4GYV5) fused to three tandem copies of the HA epitope (YPYDVPDYAGSYPYDVPDYAGSYPYDVPDYA) at its carboxyl terminus.
Protocol: The U-ExM protocol was described previously (Gorilak et al., 2021). Briefly, live trypanosomes growing in SDM-79 medium (Life Technologies #07490916 N) at a cell density between 5 × 106 and 1 × 107 cells/mL were centrifuged at 800g in a tabletop microcentrifuge. The pellet containing live trypanosome cells was washed once in vPBS buffer (137 mM NaCl, 3 mM KCl, 16 mM Na₂HPO₄, 3 mM KH₂PO₄, 46 mM sucrose) and resuspended at a final density of 4 × 106 cells in 50 μL of PBS. Resuspended cells were settled onto a 12 mm diameter glass coverslip for 10 minutes. Detergent-extracted “cytoskeletons” were prepared by aspirating excess liquid and pipetting 50 μL of 0.5% IGEPAL (Sigma-Aldrich #I8896) in PEME buffer (100 mM PIPES–NaOH pH 6.9, 1 mM MgSO₄, 2 mM EGTA, 0.1 mM EDTA) onto the coverslip, followed by incubation for 1 minute at room temperature. For fixation, the excess buffer was aspirated, and the cytoskeletons were washed twice with 50 μL of PEME. Subsequently, 500 μL of PBS was added, and the samples were incubated overnight in 500 μL of freshly prepared fixative containing 0.7% formaldehyde (Clinisciences #15710) and 1% acrylamide (Sigma-Aldrich #A4058) in PBS at room temperature in the dark.For gelation, the fixative was aspirated, and the coverslips were washed in PBS and placed cell-side-down on a 50 μL drop of monomer solution containing 19% sodium acrylate, 10% acrylamide, and 0.1% N,N'-methylenebisacrylamide (2% solution, Sigma-Aldrich #M1533) in PBS. This solution was freshly supplemented with 0.5% TEMED (Tetramethylethylenediamine, Sigma-Aldrich #T9281) and 0.5% APS (ammonium persulfate, ThermoScientific #11816714) on parafilm on ice inside a humidity chamber for 5 minutes. The gel was then transferred to a 37°C humidity chamber for 30 minutes and boiled in denaturation buffer (50 mM Tris, 200 mM NaCl, 200 mM SDS, pH 9) for 90 minutes. The gel was then expanded with three 30-minute washes in 15 mL of distilled H₂O and cut into 2 cm × 2 cm squares for antibody staining.The gel was blocked in antibody diluent (2% BSA, 0.2% Tween 20 in PBS) for at least 30 minutes and then stained with the anti-HA antibody diluted 1/500 (0.2 μg/mL final concentration) in antibody diluent overnight at room temperature with gentle agitation. After three 20-minute washes in 15 mL distilled H₂O, the gel was stained with the secondary antibody (goat anti-human Alexa 647, Invitrogen #A48279) in antibody diluent for at least 6 hours. The gel was then counterstained with guinea pig anti-tubulin (AA345) and anti-guinea pig secondary antibody (Goat anti-guinea pig Alexa 555, Invitrogen #A21435) as described above. Gels were imaged on poly-L-lysine (Sigma-Aldrich #P4707) treated glass-bottom petri dishes (Avator #75856-742) using a Leica DMi8 fluorescence microscope with a 100x 1.32 NA objective (506526), a metal halide light source, and an ORCA-Flash 4.0 sCMOS camera (Hamamatsu).
Results and discussion
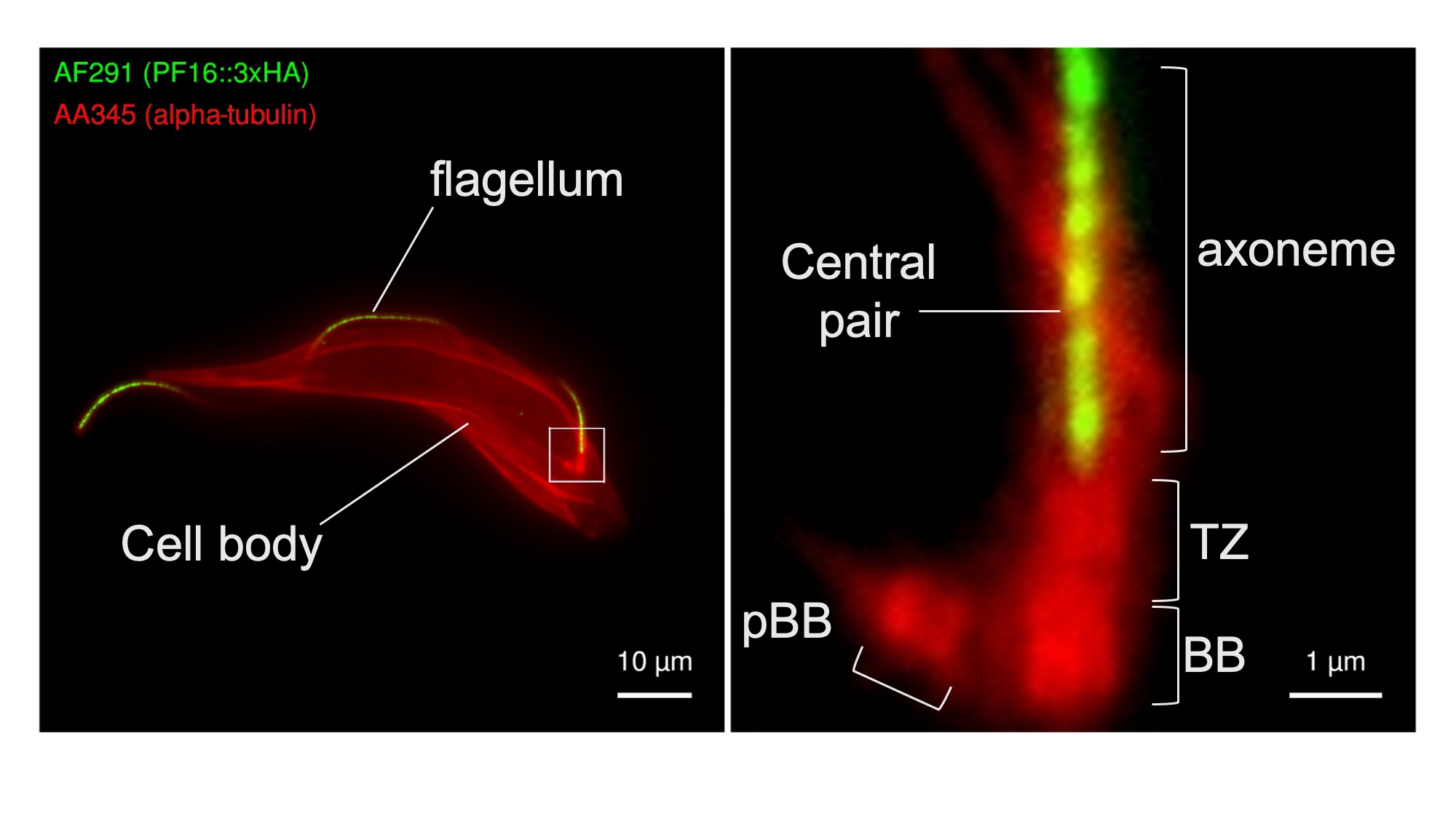
Procyclic form trypanosomes were genetically modified to express PF16 fused to three copies of the HA epitope in tandem repeat (PF16::3xHA) and subjected to U-ExM analysis using the anti-HA antibody AF291. Importantly, PF16 was tagged at its endogenous locus without using an ectopic promoter to drive its expression and is therefore expected to be expressed at physiological levels. Cells were post-stained using the anti-alpha tubulin antibody AA345 to provide informative cellular context for anti-HA staining. Consistent with the expected localisation of PF16, anti-HA staining produced a strong signal in the centre of the flagellar axoneme that emanated from its base, with only background signal elsewhere. Anti-HA staining gave only background signal when untransfected control cells were stained (data not shown). This demonstrates that AF291 can be used for U-ExM detection of HA-tagged trypanosome proteins. Further, the availability of engineered AF291 variants that differ only in their Fc domain will facilitate flexible co-staining and co-localisation analyses.
Figure 1. The AF291 antibody recognizes the HA epitope in U-ExM of detergent extracted T. brucei cytoskeletons. Transgenic T. brucei cells expressing PF16::3HA were analyzed by U-ExM using the anti-HA antibody AF291 (green), and counterstained with the AA345 anti-alpha tubulin (red), to provide cellular context. Images were captured on a Leica DMi8; projected z-stacks of the whole cell signal are shown (left). Consistent with a flagellar central pair localization, the anti-HA antibody signal runs along the cell body of the parasite. The magnified inset (right) shows the base of the flagellum with the anti-HA signal emerging from the axoneme base. Key structural features of the flagellar base are marked: pro-basal body (pBB), basal body (BB), and flagellar transition zone (TZ).
Conflict of interest
The authors declare no conflict of interest.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgements
AP was supported by an Academy of Medical Sciences Springboard award [SBF006/1126] and BBSRC NIRG (BB/Y012739/1) to SD. JSC is supported by a doctoral scholarship from the Warwick-A*STAR research attachment program and SN was supported by an MRC doctoral training program award. We would like to thank Vladimir Varga (Czech Academy of Sciences) for his help establishing U-ExM in our laboratory and the Warwick Medical School for their institutional support.
Author contributions
AP performed the U-ExM and helped write the manuscript, JSC and SN helped establish the method, SD provided funding, supervision, and wrote the manuscript.
References
Brun, R., & Schönenberger, null. (1979). Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Tropica, 36(3), 289–292.
Dean, S., Sunter, J., Wheeler, R. J., Hodkinson, I., Gluenz, E., & Gull, K. (2015). A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biology, 5(1), 140197. https://doi.org/10.1098/rsob.140197
Gorilak, P., Pružincová, M., Vachova, H., Olšinová, M., Schmidt Cernohorska, M., & Varga, V. (2021). Expansion microscopy facilitates quantitative super-resolution studies of cytoskeletal structures in kinetoplastid parasites. Open Biology, 11(9), 210131. https://doi.org/10.1098/rsob.210131
Paterou, A., Týč, J., Sunter, J., Vaughan, S., Gull, K., & Dean, S. (2023). Unlocking Trypanosome Biology: A Comprehensive Protein-Tagging Toolkit for Localization and Functional Analysis. BioRxiv. https://doi.org/10.1101/2023.04.21.537815
Poon, S. K., Peacock, L., Gibson, W., Gull, K., & Kelly, S. (2012). A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biology, 2(2), 110037. https://doi.org/10.1098/rsob.110037
Smith, E. F., & Lefebvre, P. A. (1996). PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. The Journal of Cell Biology, 132(3), 359–370. https://doi.org/10.1083/jcb.132.3.359
Downloads
Published
Section
How to Cite
License
Some rights reserved 2024 Athina Paterou, Julia Sáez Conde, Shambhawee Neupane, Samuel Dean

This work is licensed under a Creative Commons Attribution 4.0 International License.



